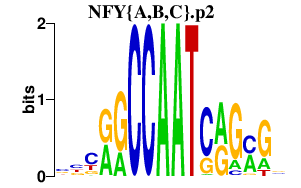
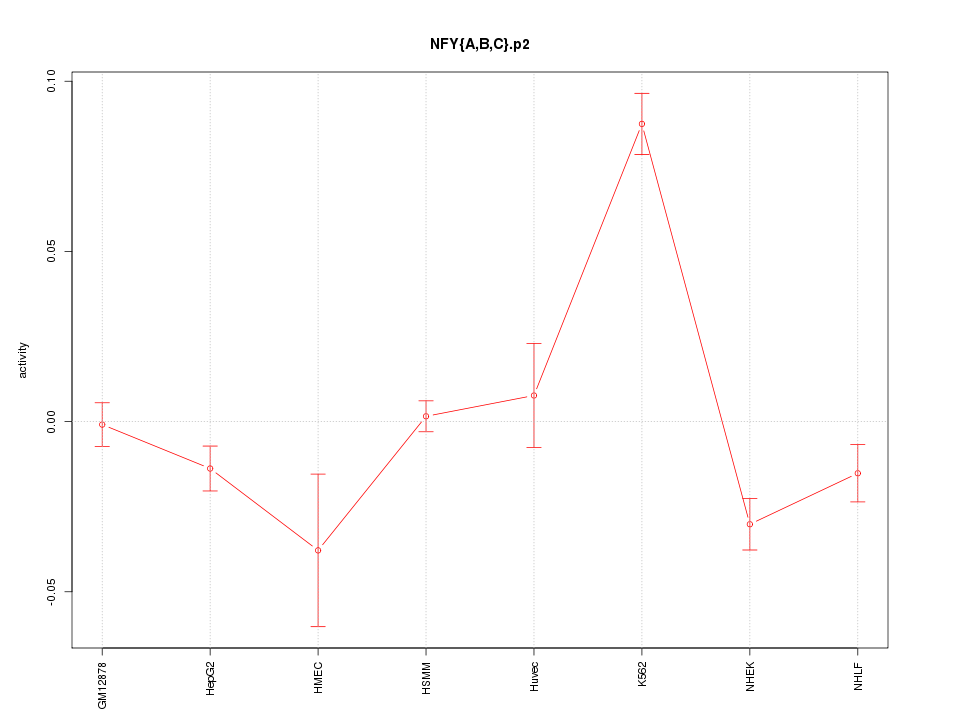
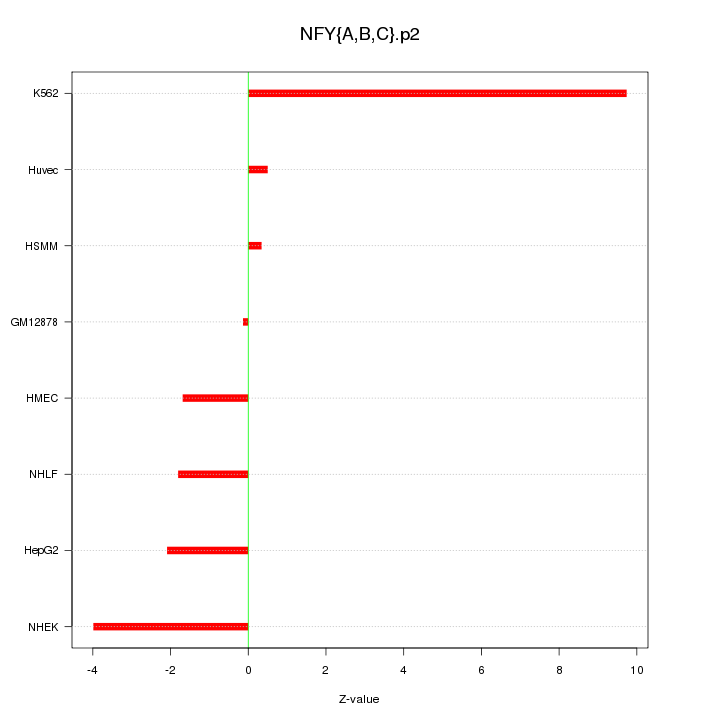
|
chr20_-_538909
|
7.433
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr17_+_54587641
|
5.312
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr17_-_77411733
|
5.108
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr17_-_54587581
|
5.001
|
NM_001100595
NM_182620
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr11_+_61473931
|
4.844
|
NM_001139443
NM_004183
|
BEST1
|
bestrophin 1
|
|
chr17_-_77411695
|
4.751
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chrX_+_47999740
|
4.493
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr17_-_77411659
|
4.490
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr7_-_150305933
|
4.456
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr17_-_77411707
|
4.333
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr17_-_77411803
|
4.114
|
NM_000918
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr21_+_44761927
|
4.016
|
|
C21orf90
|
chromosome 21 open reading frame 90
|
|
chr1_+_43888391
|
3.990
|
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr3_-_121295663
|
3.923
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr7_-_71515295
|
3.899
|
NM_031468
|
CALN1
|
calneuron 1
|
|
chr1_+_43888415
|
3.832
|
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr3_+_9414290
|
3.819
|
NM_001080517
|
SETD5
|
SET domain containing 5
|
|
chr17_+_54587882
|
3.723
|
|
PRR11
|
proline rich 11
|
|
chr1_+_43888382
|
3.688
|
NM_014663
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr6_-_25834733
|
3.684
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr17_-_77411607
|
3.614
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr1_+_110683580
|
3.486
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr1_+_110683586
|
3.471
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr22_-_21193037
|
3.427
|
|
ZNF280B
|
zinc finger protein 280B
|
|
chr15_-_97368546
|
3.399
|
NM_001167902
|
PGPEP1L
|
pyroglutamyl-peptidase I-like
|
|
chr19_+_46462113
|
3.387
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr6_+_25834983
|
3.376
|
NM_170610
|
HIST1H2BA
|
histone cluster 1, H2ba
|
|
chr1_-_159257486
|
3.322
|
|
F11R
|
F11 receptor
|
|
chr1_+_110683467
|
3.140
|
NM_022768
|
RBM15
|
RNA binding motif protein 15
|
|
chr19_+_45794895
|
3.081
|
NM_001042544
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr22_-_20419975
|
3.034
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr19_-_54350457
|
3.020
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr19_+_46462125
|
3.003
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr1_-_148868713
|
2.975
|
NM_004436
NM_207042
NM_207043
NM_207044
NM_207168
|
ENSA
|
endosulfine alpha
|
|
chr19_+_46462176
|
2.975
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr5_-_95323180
|
2.932
|
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr7_+_129497637
|
2.918
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr7_+_129497572
|
2.840
|
NM_014997
|
KLHDC10
|
kelch domain containing 10
|
|
chr19_+_46462169
|
2.838
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr9_-_73715666
|
2.832
|
|
FAM108B1
|
family with sequence similarity 108, member B1
|
|
chrX_+_22927998
|
2.818
|
NM_182699
|
DDX53
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 53
|
|
chr9_-_127043436
|
2.815
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr16_+_1772929
|
2.783
|
NM_012225
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr19_-_6375801
|
2.744
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr9_-_127043455
|
2.735
|
NM_005347
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr19_+_46462189
|
2.721
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr1_-_27689129
|
2.702
|
|
WASF2
|
WAS protein family, member 2
|
|
chr9_-_127043169
|
2.701
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr1_-_148868671
|
2.695
|
|
ENSA
|
endosulfine alpha
|
|
chr7_+_98940208
|
2.665
|
NM_145102
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr14_+_28305922
|
2.661
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr11_-_64302440
|
2.652
|
|
SF1
|
splicing factor 1
|
|
chr18_-_43711143
|
2.624
|
|
SMAD2
|
SMAD family member 2
|
|
chr7_-_73944616
|
2.596
|
|
STAG3L2
|
stromal antigen 3-like 2
|
|
chr1_-_27689254
|
2.585
|
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr19_-_12858956
|
2.562
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr1_+_28458546
|
2.554
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr3_+_9414533
|
2.541
|
|
SETD5
|
SET domain containing 5
|
|
chr1_-_180627512
|
2.532
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr4_+_166468266
|
2.528
|
NM_001017369
NM_006745
|
SC4MOL
|
sterol-C4-methyl oxidase-like
|
|
chr16_+_1773004
|
2.520
|
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr17_-_44037332
|
2.506
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr7_+_31523502
|
2.449
|
NM_194300
|
CCDC129
|
coiled-coil domain containing 129
|
|
chr7_-_137337318
|
2.440
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_-_109742020
|
2.434
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chrX_-_77037563
|
2.425
|
NM_032121
|
MAGT1
|
magnesium transporter 1
|
|
chrX_+_52797004
|
2.424
|
NM_003147
NM_175698
NM_001164417
|
SSX2
SSX2B
|
synovial sarcoma, X breakpoint 2
synovial sarcoma, X breakpoint 2B
|
|
chr7_+_129497808
|
2.413
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr7_+_115637795
|
2.410
|
NM_015641
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr7_-_137337352
|
2.402
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_-_159257567
|
2.342
|
|
F11R
|
F11 receptor
|
|
chr14_-_99016959
|
2.319
|
NM_032233
NM_199123
|
SETD3
|
SET domain containing 3
|
|
chr8_+_11697588
|
2.317
|
NM_004462
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr17_-_54587306
|
2.302
|
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr7_-_150306169
|
2.290
|
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr21_+_39745649
|
2.286
|
NM_007341
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr22_+_20316968
|
2.267
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr4_-_174492029
|
2.263
|
NM_001130688
NM_002129
|
HMGB2
|
high-mobility group box 2
|
|
chr19_+_4920123
|
2.262
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr3_-_9414122
|
2.246
|
|
LOC440944
|
hypothetical LOC440944
|
|
chr19_-_1992862
|
2.239
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr17_+_71772982
|
2.229
|
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr7_+_74826369
|
2.229
|
NM_001002840
NM_018991
|
STAG3L3
STAG3L2
STAG3L1
|
stromal antigen 3-like 3
stromal antigen 3-like 2
stromal antigen 3-like 1
|
|
chr14_-_99016917
|
2.220
|
|
SETD3
|
SET domain containing 3
|
|
chr3_+_37009987
|
2.213
|
|
MLH1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli)
|
|
chr4_+_78297418
|
2.212
|
|
CCNG2
|
cyclin G2
|
|
chr16_-_57221235
|
2.203
|
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr22_-_21193504
|
2.189
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr12_-_54497751
|
2.165
|
|
SARNP
|
SAP domain containing ribonucleoprotein
|
|
chr2_-_61618898
|
2.145
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr11_+_67984758
|
2.142
|
NM_001164160
NM_001164161
NM_001164162
NM_001164163
NM_001164164
NM_018312
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr16_-_1816793
|
2.136
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr11_+_58103212
|
2.132
|
|
ZFP91-CNTF
ZFP91
|
ZFP91-CNTF readthrough transcript
zinc finger protein 91 homolog (mouse)
|
|
chrX_-_100549561
|
2.103
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr1_-_35431017
|
2.097
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr16_-_1772577
|
2.095
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr11_+_82545784
|
2.093
|
NM_015885
|
PCF11
|
PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae)
|
|
chrX_-_153016368
|
2.087
|
NM_001110792
NM_004992
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome)
|
|
chr1_+_24158887
|
2.082
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr5_+_44844724
|
2.082
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr3_-_37009613
|
2.078
|
NM_014805
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chr1_+_42920755
|
2.071
|
|
YBX1
|
Y box binding protein 1
|
|
chr1_+_153545301
|
2.069
|
NM_001135821
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr1_+_42920729
|
2.066
|
|
YBX1
|
Y box binding protein 1
|
|
chrX_+_46889563
|
2.054
|
NM_005676
NM_152856
|
RBM10
|
RNA binding motif protein 10
|
|
chr1_-_148868664
|
2.044
|
|
ENSA
|
endosulfine alpha
|
|
chr1_+_5975286
|
2.041
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr1_-_148868644
|
2.025
|
|
ENSA
|
endosulfine alpha
|
|
chr1_+_42920774
|
2.022
|
|
YBX1
|
Y box binding protein 1
|
|
chr11_-_64302509
|
2.001
|
NM_001178030
|
SF1
|
splicing factor 1
|
|
chr3_-_41978464
|
1.999
|
NM_017886
|
ULK4
|
unc-51-like kinase 4 (C. elegans)
|
|
chr19_+_55398680
|
1.960
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr19_+_62434188
|
1.958
|
NM_001015879
NM_003160
NM_001015878
|
AURKC
|
aurora kinase C
|
|
chr17_-_71287417
|
1.952
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr3_-_37009592
|
1.949
|
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chr22_-_22511173
|
1.947
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chrX_-_153016337
|
1.945
|
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome)
|
|
chr12_+_53005177
|
1.945
|
NM_016057
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr7_-_100092003
|
1.944
|
NM_016188
|
ACTL6B
|
actin-like 6B
|
|
chr1_-_233558114
|
1.944
|
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr1_-_233558071
|
1.943
|
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr20_+_47096280
|
1.940
|
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr22_-_20551937
|
1.939
|
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr21_-_33836994
|
1.939
|
NM_001136005
NM_001136006
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr22_-_20551953
|
1.938
|
NM_002745
NM_138957
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr19_-_6375804
|
1.938
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr7_+_149707338
|
1.933
|
NM_173680
|
ZNF775
|
zinc finger protein 775
|
|
chr1_-_159257595
|
1.929
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr7_-_100091912
|
1.927
|
|
ACTL6B
|
actin-like 6B
|
|
chr1_+_159741824
|
1.925
|
NM_001136219
NM_021642
|
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32)
|
|
chr3_-_187771025
|
1.918
|
NM_001134415
|
TBCCD1
|
TBCC domain containing 1
|
|
chr16_-_1816529
|
1.917
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr3_+_37009844
|
1.916
|
NM_000249
|
MLH1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli)
|
|
chr21_-_39642712
|
1.913
|
|
HMGN1
|
high-mobility group nucleosome binding domain 1
|
|
chr6_+_31891293
|
1.909
|
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr1_-_219982016
|
1.907
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr12_+_53005183
|
1.907
|
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr1_-_219982049
|
1.906
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr17_+_71772875
|
1.903
|
NM_182565
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chrX_-_47394808
|
1.900
|
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr7_-_135063461
|
1.898
|
NM_012450
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporters), member 4
|
|
chr9_-_73715927
|
1.898
|
NM_001025780
NM_016014
|
FAM108B1
|
family with sequence similarity 108, member B1
|
|
chr1_-_233558117
|
1.891
|
NM_016374
NM_031371
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr1_+_153545351
|
1.883
|
|
FDPS
|
farnesyl diphosphate synthase
|
|
chrX_-_47394937
|
1.883
|
NM_001114123
NM_005229
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr16_-_1772572
|
1.879
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr16_-_57221228
|
1.877
|
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr3_+_187771139
|
1.873
|
NM_016306
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11
|
|
chr21_-_39642880
|
1.872
|
NM_004965
|
HMGN1
|
high-mobility group nucleosome binding domain 1
|
|
chrX_+_46889601
|
1.865
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr21_-_33774033
|
1.862
|
|
TMEM50B
|
transmembrane protein 50B
|
|
chr16_-_57221240
|
1.858
|
NM_016284
NM_206999
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr3_-_187770790
|
1.849
|
|
TBCCD1
|
TBCC domain containing 1
|
|
chr1_+_42920718
|
1.846
|
|
YBX1
|
Y box binding protein 1
|
|
chrX_+_46889620
|
1.844
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr1_-_148868535
|
1.842
|
|
ENSA
|
endosulfine alpha
|
|
chr1_+_25629968
|
1.838
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr21_-_39642935
|
1.838
|
|
HMGN1
|
high-mobility group nucleosome binding domain 1
|
|
chr7_-_112514990
|
1.837
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr7_-_112515035
|
1.836
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chrX_+_46889614
|
1.828
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr12_+_53005192
|
1.824
|
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr7_-_112515068
|
1.820
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr14_+_57835028
|
1.815
|
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr14_-_22458220
|
1.813
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr17_+_71773026
|
1.811
|
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr21_+_33837215
|
1.810
|
NM_032195
NM_138927
|
SON
|
SON DNA binding protein
|
|
chr3_-_37009571
|
1.807
|
|
|
|
|
chr1_+_24158898
|
1.802
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr21_+_33837212
|
1.784
|
|
SON
|
SON DNA binding protein
|
|
chr2_-_130672481
|
1.784
|
NM_207312
|
TUBA3E
|
tubulin, alpha 3e
|
|
chr19_-_6375820
|
1.784
|
NM_003685
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chrX_+_100549869
|
1.780
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_+_46889609
|
1.772
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr8_-_25371708
|
1.769
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chrX_+_100549839
|
1.763
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_-_52700674
|
1.763
|
NM_173358
|
SSX7
|
synovial sarcoma, X breakpoint 7
|
|
chr8_-_25371858
|
1.763
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr15_+_81275383
|
1.762
|
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chr19_-_6375782
|
1.762
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr7_-_73944644
|
1.760
|
NM_001025202
|
STAG3L2
|
stromal antigen 3-like 2
|
|
chr14_+_57834962
|
1.758
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr5_+_145807059
|
1.753
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr8_-_103737058
|
1.738
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr19_-_1993022
|
1.735
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr1_+_42920889
|
1.732
|
|
YBX1
|
Y box binding protein 1
|
|
chr1_-_28113817
|
1.731
|
NM_002946
|
RPA2
|
replication protein A2, 32kDa
|
|
chrX_+_46889577
|
1.727
|
|
RBM10
|
RNA binding motif protein 10
|
|
chrX_-_153016173
|
1.721
|
|
MECP2
LOC728653
|
methyl CpG binding protein 2 (Rett syndrome)
hypothetical protein LOC728653
|
|
chr22_-_20551722
|
1.721
|
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr9_+_132990834
|
1.712
|
|
NUP214
|
nucleoporin 214kDa
|
|
chr19_-_17377383
|
1.709
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chrX_+_46889637
|
1.707
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr1_-_151910080
|
1.702
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr11_+_63710242
|
1.698
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chrX_+_100549861
|
1.696
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_-_35431318
|
1.693
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr5_+_177564140
|
1.692
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr1_+_153545162
|
1.691
|
NM_001135822
NM_002004
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr1_+_165225121
|
1.684
|
NM_032858
|
MAEL
|
maelstrom homolog (Drosophila)
|